** At the end of this page, you can find the full list of publications (peer-reviewed and preprints).

GBPs are dynamin-like GTPases that act as core orchestrators of the cell-autonomous immune response by coating the membranes of intracellular pathogen replication niches and/or cytosol-invasive gram-negative bacteria. We used cryo-EM/cryo-ET to resolve the molecular ultrastructure of GBP1 coatomers and reveal the large-scale conformational changes that allow GBP1 to selectively target LPS-containing membranes. We further demonstrate, for the first time, that GTP hydrolysis drives GBP1-dependent membrane fragmentation, which possibly underlies the ability to recruit and activate caspase4-dependent inflammasomes.
Kuhm T, Taisne CM, de Agrela Pinto C, Gross L, Giannopolou EA, Huber ST, Pardon E, Steyaert J, Tans S, Jakobi AJ
Nature Structural & Molecular Biology 32:172–184 (2025)
PDBe: 8cqb
EMDB: EMD-16813, EMD-16814, EMD-16815, EMD-16794
Featured as NSMB Research Briefing.
Highlighted in the microbiologist, TU Delft News, phys.org.

Gas vesicles are nanoscale buoyancy compartments that form a unique motility system in (aquatic) bacteria and archaea. Using cryo-EM, we reveal the first molecular structure of the intact gas vesicle shell at 3.2 Å resolution. Our structure explains the molecular details underlying their unique properties such as the selective permeability of the shell and the role of structural reinforcement elements, consolidating decades of biochemical and biophysical studies. Our structure also allows to formulate a testable hypothesis of the intrigiuing structural program of gas vesicle assembly and growth.
Huber ST, Terwiel D, Evers W, Maresca D#, Jakobi AJ#
PDBe: 7r1c
EMDB: EMD-14238
Featured as News & Views in Nature Structural & Molecular Biology by Katarzyna Ciazynska.
Highlighted in TU Delft News and phys.org
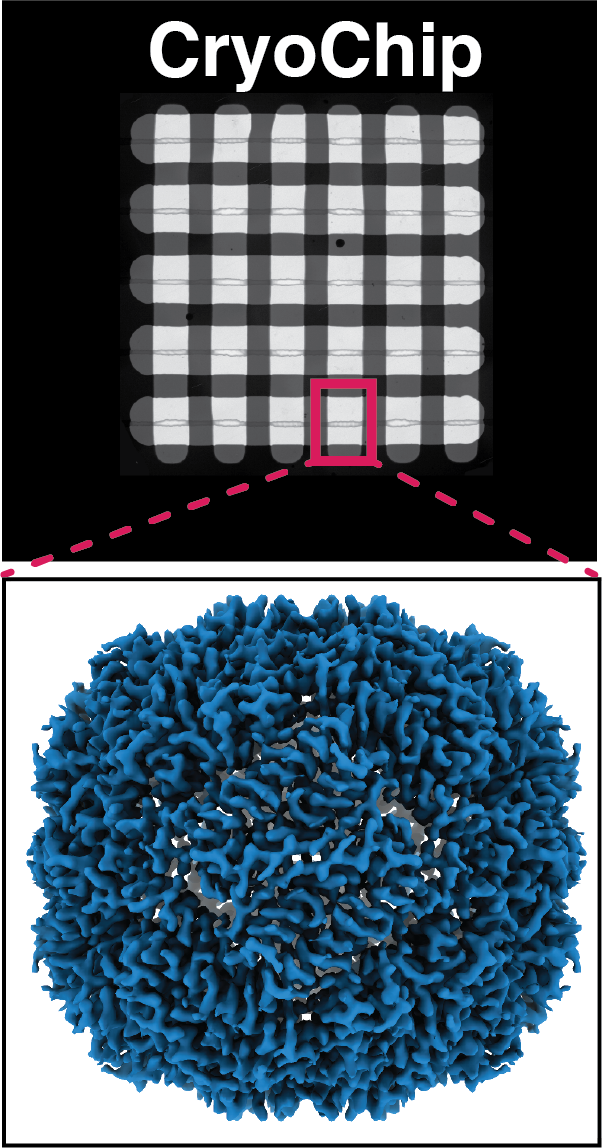
We have developed a microelectromechanical system (MEMS) in which macromolecular targets are enclosed in nanofluidic channels formed between ultrathin, electron-transparent membranes. We show that these devices are suitable for high-resolution 3D structure determination of macromolecular complexes from picoliter sample volumes. Our device protects the sample from a destructive air-water interface and provides excellent control over geometric parameters of the nanochannels which yields uniform ice thickness.
Huber ST, Sarajilic E, Huijink R, Evers WH, Jakobi AJ
PDB: 7ohf
EMDB: EMD-12901, EMD-12902, EMD-12903, EMD-12914, EMD-12915, EMD-12917
Methods to improve interpretation of electrostatic potential maps from cryo-EM reconstructions are essential to build accurate atomic models. For all downstream applications it is important that any form of manipulation of these maps retains the physical properties expected from scattering of biological macromolecules. In this article we provide a roadmap to utilising such information to restrain the process of map sharpening and discuss its potential and limitations.
Bharadwaj A, Jakobi AJ
Faraday Discussions 11:e72629 (2022)
PDBe: 6y5a
EMDB: EMD-10692
This article is discussed in the companion General Discussion of the RSC Faraday Meeting.
Associated & Supporting data EMPIAR data Associated code Associated movies
Confidence-guided cryo-EM map optimisation with LocScale-2.0
Bharadwaj A, de Bruin RMM, Jakobi AJ
BioRxiv 2025.09.11.674726 (2025)
– this article is a BioRxiv preprint and has not yet been certified by peer review.
Femtolitre volume cryo-EM sample preparation using a force-sensitive microfluidic cantilever
Shastri V†, Pronk J†, Verlinden E†, Torres Gonzàles D, Sarajilic E, Laeven P, Evers WH, Frederix P, Staufer U, Jakobi AJ#, Ghatkesar MK#
Workflow for fluorescence-targeted lamella milling from vitrified cells with a coincident fluorescence, electron, and ion beam microscope
Perton EG, Boltje DB, Jakobi AJ, Hoogenboom JP
Protocol associated with
Boltje DB et al.: A cryogenic, coincident fluorescence, electron, and ion beam microscope eLife 11:e82891 (2022)
Thickness and quality controlled fabrication of fluorescence-targeted frozen-hydrated lamellae
Boltje DB, Skoupý R, Taisne CM, Evers WH, Jakobi AJ, Hoogenboom JP
Enterovirus-like particles encapsidate RNA and exhibit decreased stability due to lack of maturation
Kuijpers L, Giannopoulou EA, Feng Y, van den Braak W, Freydoonian A, Ramlal R, Meiring H, Solano B, Roos W, Jakobi AJ, van der Pool LA, Dekker NH
Snapshots of Pseudomonas aeruginosa SOS response activation complex reveal structural prerequisites for LexA engagement and cleavage
Vascon F†, De Felice S†, Gasparotto M, Huber S, Catalano C, Chinellato M, Grinzato A, Filipini F, Maso M, Jakobi AJ, Cendron L
PDBe: 8b0v, 8s70, 8s7g
EMDB: EMD-19761, EMD-19771
Structural basis of antimicrobial membrane coat assembly by human GBP1
Kuhm T, Taisne CM, de Agrela Pinto C, Gross L, Giannopolou EA, Huber ST, Pardon E, Steyaert J, Tans S, Jakobi AJ
Nature Structural & Molecular Biology 32:172–184 (2025)
PDBe: 8cqb
EMDB: EMD-16813, EMD-16814, EMD-16815, EMD-16794
Featured as NSMB Research Briefing.
The guanylate-binding protein GBP1 forms a protein coat that enwraps cytosol-invasive bacteria
Jakobi AJ
Nature Structural & Molecular Biology 32:8–9 (2025)
Research Briefing associated with Kuhm et al.:Structural basis of antimicrobial membrane coat assembly by human GBP1.
Roodmus: A toolkit for benchmarking heterogeneous electron cryo-microscopy reconstructions
Joosten M†, Greer J†, Parkhurst J, Burnley T#, Jakobi AJ#
Roodmus is a Python-based toolkit to generate synthetic cryo-EM datasets of continous molecular heterogeneity by sourcing conformational variability from molecular dynamics simulations. Its development was a joint effort with Joel Greer and Tom Burnley at CCPEM.
Structure-based mechanism of riboregulation of the metabolic enzyme SHMT1
Spizzichino S, Di Fonzo F, Marabelli C, Tramonti A, Chaves-Sanjuan A, Parroni A, Boumis G, Romano Liberati F, Paone A, Montemiglio LC, Ardini M, Jakobi AJ, Bharadwaj A, Tartaglia GG, Paiardini A, Contestabile R, Swuec P, Rinaldo S, Bolognesi M, Giardina G, Cutruzzolà F
Molecular Cell 84:2682-2697.e6 (2024)
PDBe: 8r7h, 8a11, 8s7g
EMDB: EMD-18973, EMD-15065
Structural biology of microbial gas vesicles: historical milestones and current knowledge
Huber ST and Jakobi AJ
Biochemical Society Transactions 52:205-215 (2024)
In this review article we review the 60-year journey in quest for a high-resolution structural model of gas vesicles. Stefan’s beautiful illustration was featured on the cover!
Particle fusion of super-resolution data reveals the unit structure of Nup96 in Nuclear Pore Complex
Wang W, Jakobi AJ, Wu Y-L, Ries J, Stallinga S, Rieger B
Scientific Reports 13:13327 (2023)
A publisher correction is associated with this article.
Robust local thickness estimation of sub-micrometer specimen by 4D-STEM
Skoupý R, Boltje DB, Slouf M, Mrázová K, Láznička T, Taisne CM, Krzyzanek, Hoogenboom JP, Jakobi AJ
Cryo-EM structure of gas vesicles for buoyancy-controlled motility
Huber ST, Terwiel D, Evers W, Maresca D#, Jakobi AJ#
PDBe: 7r1c
EMDB: EMD-14238
Featured as News & Views in Nature Structural & Molecular Biology by Katarzyna Ciazynska.
Multivalent interactions facilitate motor-dependent protein accumulation at growing microtubule plus ends
Maan R, Reese L, Volkov VA, King MR, van der Sluis E, Andrea N, Evers WH, Jakobi AJ, Dogterom M
Nature Cell Biol. 25:975-986 (2023)
EMDB: EMD-14106, EMD-14107, EMD-14108, EMD-14109, EMD-14110, EMD-14110, EMD-14113
A cryogenic, coincident fluorescence, electron, and ion beam microscope
Boltje DB, Hoogenboom JP, Jakobi AJ, Jensen GJ, Jonker CTH, Kaag MJ, Koster AJ, Last MGF, de Agrela Pinto C, Plitzko JM, Raunser S, Tacke S, Wang Z, van der Wee EB, Wepf R, ten Hoedt S
Highlighted in TU Delft News
Electron scattering properties of biological macromolecules and their use in cryo-EM map sharpening
Bharadwaj A, Jakobi AJ
Map/model validation and machine learning in EM
Bakker SE, Bergeron J, Bharadwaj A, Bhella D, Bullough P, Chau P-L, Frank RAW, Jakobi AJ, Joseph AP, Kühlbrandt W, Lahiri I, Menday R, Muench SP, Nakasone M, Nerukh D, Paris G, Russo CJ, Saibil HR, Scheres SHW, Sherawat V, Shah AR, Thorn A, Vilas JL, Zanetti G
Faraday Discussion 240:229-242 (2022)
This is a companion article to the above paper containing the public discussion of the paper at the Faraday Discussion “Challenges in biological cryo electron microscopy Faraday Discussion”
Guidelines for the use and interpretation of assays for monitoring autophagy
Klionsky D, Abdel-Aziz AK, Abdelfatah S, Abdoli A, Abel S, …, Jakobi AJ, et.al.
Structural basis of p62/SQSTM1 helical filaments and their role in cellular cargo uptake
Jakobi AJ, Huber ST, Mortensen SA, Schultz SW, Palara A, Kuhm T, Shrestha BK, Lamark T, Johansen T, Brech A, Sachse C
Nature Communications 11:440 (2020)
PDB: 6tgn, 6tgp, 6tgs, 6th3, 6tgy
EMDB: EMD-10499, EMD-10500, EMD-10501, EMD-10502,

Seeing with electrons
Arjen J. Jakobi
Europhysics News 51:20-23 (2020)
This short overview article is part of the EPN Special Issue on Physics of Living Matter
Improvement of production and isolation of human neuraminidase-1 in cellulo crystals
Koiwai K, Tsukimoto J, Higashi T, Mafune F, Miyajima K, Nakane T, Matsugaki N, Jakobi AJ, Sugahara M, Tanaka T, Tono K, Joti Y, Yabashi M, Nureki O, Mizohata E, Nakatsu T, Nango E, Iwata S, Chavas LMG, Senda T, Itoh K, Yumoto F
Thresholding of cryo-EM density maps by false discovery rate control]
Beckers M, Jakobi AJ, Sachse C
Highlighted in IUCrJ Editorial Notes
An atomic model of the HIV-1 capsid-SP1 layer reveals structures regulating assembly and maturation
Schur FKM, Obr M, Wan W, Jakobi AJ, Sachse C, Krausslich HG, Briggs JAG
Characterization of Atg38 and NRBF2, a fifth subunit of the autophagic Vps34 complex
Ohashi Y, Soler N, Garcia Ortegon M, Zhang L, Kirsten M, Perisic O, Masson, GR, Burke J, Jakobi AJ, Apostolakis AA, Johnson CM, Ohashi M, Ktistakis NT, Sachse C, Williams RL
Higher-order assemblies of oligomeric cargo receptor complexes form the membrane scaffold of the Cvt vesicle
Bertipaglia C, Jakobi AJ, Tarafder AK, Bykov YS, Picco A, Kukulski W, Kosinski J, Hagen WJH, Wilmanns M, Kaksonen M, Briggs JAG, Sachse C
EMBO Reports 17:1044-1060 (2016)
PDB: 5jm0, 5jm6
EMDB: EMD-8166
Featured in EMBOpress Collection on Autophagy and highlighted in EMBL News
Transcribing RNA polymerase III observed by electron cryo-microscopy
Hoffmann NA, Jakobi AJ, Vorlander MK, Sachse C, Muller CW
In cellulo X-ray Diffraction of Native Alcohol Oxidase Crystals in Yeast Cells by Serial Femtosecond Crystallography
Jakobi AJ#, Passon DM, Knoops K, Stellato F, White TA, Seine T, Messerschmidt M, Chapman HN, Wilmanns M
Highlighted in IUCrJ Editorial Notes, DESY Photon Science Highlights, DESY News, Femto, Research in Germany, CUI, chemie.de
An organized co-assembly of clathrin adaptors is essential for endocytosis
Skruzny M, Desfosses A, Prinz S, Dodonova S, Uetrecht C, Gieras A, Jakobi AJ, Abella M, Hagen WJH, Schulz J, Meijers R, Rybin V, Briggs JAG, Sachse C, Kaksonen M
Developmental Cell 33:150-162 (2015)
PDB: 5ahv
EMDB: EMD-2896, EMDB: EMD-2897
Highlighted in EMBL News
Characterization of the mycobacterial Acyl-CoA carboxylase holo complexes reveals their functional expansion into amino acid catabolism
Ehebauer MT, Jakobi AJ, Noens EE, Laubitz D, Cichocki B, Marrakchi H, Laneelle MA, Daffe M, Sachse C, Dziembowski A, Sauer U, Wilmanns M
Novel Sequence Variation affects GPIba in Post-diarrheal Hemolytic Uremic Syndrome
Volokhina E, Jakobi AJ, Urbanus RT, Huizinga EG, Sluiter H, Dineke Westra D, van Heerde W, van de Kar N, van den Heuvel L
J Nephrol Therapeutic S11:007 (2014)
The guanine nucleotide exchange factor Rlf interacts with SH3 domain-containing proteins via a binding site with a preselected conformation
Popovic M*<, Jakobi AJ*<, Rensen-de Leeuw M, Rehmann H
J Struct Biol 183:312-319 (2013)
PDB: 4jgw
Calcium modulates force sensing by the von Willebrand Factor A2 domain
Jakobi AJ, Mashaghi A, Tans S, Huizinga EG
Nature Communications 2:e385 (2012)
PDB: 3zqk
Featured in: Chemisch Weekblad C2W, NWO, AMOLF, NU.nl
A rapid cloning method employing orthogonal end protection
Jakobi AJ, Huizinga EG
An integrated fragment based screening approach for the discovery of small molecule modulators of the VWF-GPIb interaction
Jose RA, Voet A, Broos K, Jakobi AJ, Bruylants G, Egle B, Zhang KY, De Maeyer M, Deckmyn H, De Borggraeve WM
Structure of Ammonium 2-aminopyrazine-3-carboxylate
Jakobi AJ, Lutz M
Acta Cryst Section E 67:o984 (2011)
CIF: CCDC-820283
ParaFrag - a novel approach for surface-based similarity comparison of molecular fragments
Jakobi AJ, Mauser H, Clark T