We use cryo-EM to study the structures of large self-assembling macromolecular complexes, including force-generating enzyme machines that mediate intracellular immunity, protein-based compartments that impart cellular buoancy and cytoskeletal structures that organize the intracellular space. In each of these systems protein-protein interactions drive creation of large cellular structures with unique functional properties.
Our lab aims at a quantitative understanding of how these biomolecular machines function in their native biological environment. Our approach is inspired by the idea that once we understand biology at the atomic level, we can describe it with the laws of chemistry and physics to answer how its molecular components work the way they do. Biological macromolecules adopt intricate three-dimensional arrangments that are critical to their function. By taking many hundreds of thousands of images of the molecules and combining all of the information from these images, we can visualise their three-dimensional almost at atomic detail. Having a molecular picture of protein assemblies enables us to learn how they work, and provides the possibility to modify them in a way that may improve or revise their function. Since both the physical dimension and the operation level of the systems we study are at the nanoscale, we collectively describe this as biomolecular nanoscopy. We complement our structural work with functional studies using a broad array of biophysical, biochemical, and cell biological approaches.
05 July 2025
New publication: First steps towards cryo-EM sample preparation with femtoliter volumes using microfluidic cantilevers. 
10 June 2025
LocScale-SURFER is now available via the ChimeraX Toolshed 
24 May 2025
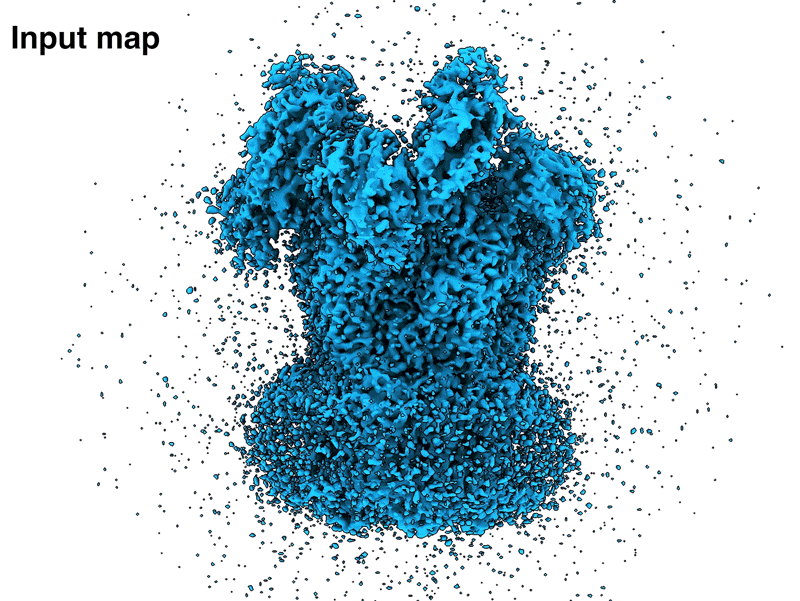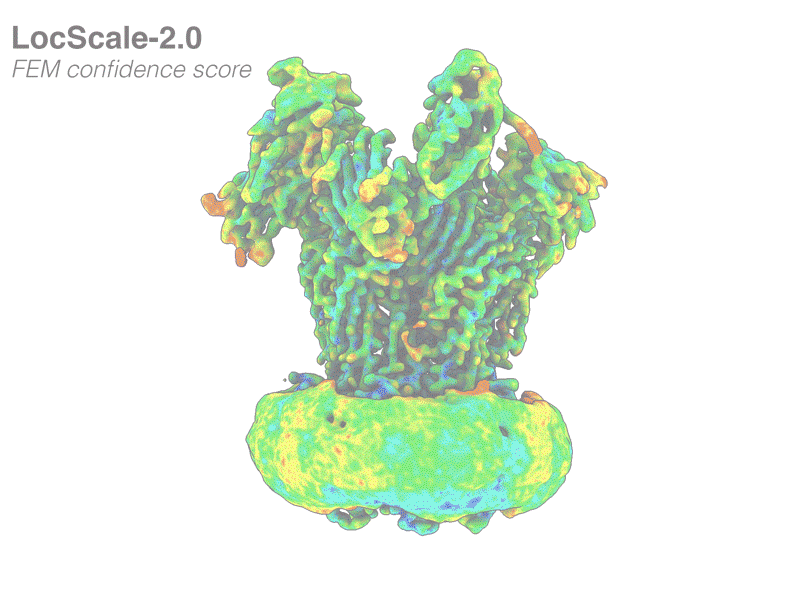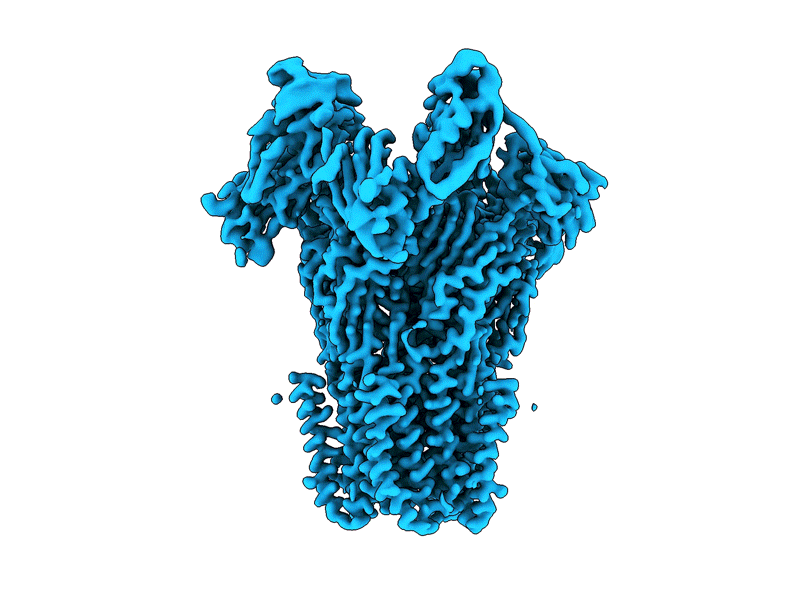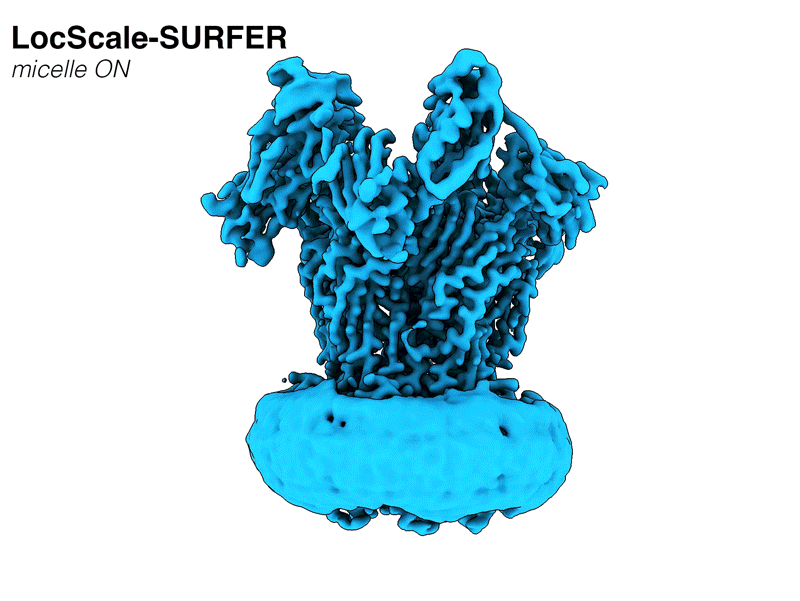
We have released LocScale-2.0, our new tool for cryoEM map optimisation. Great work by Alok, Reinier and Lotte!